Introduction#
Simulate from a Gaussian copula and use mixed-normal distributions as margins
In [1]: import numpy as np
In [2]: from scipy.stats import multivariate_normal, norm, invwishart
In [3]: from scipy.linalg import toeplitz
In [4]: from scipy.optimize import root_scalar
In [5]: class GaussianMixture:
...: def __init__(self, weights, mus, sigmas):
...: self.weights = weights
...: self.mus = mus
...: self.sigmas = sigmas
...: self.n_components = len(weights)
...: def cdf(self, x):
...: res = np.zeros_like(x)
...: for i_component in range(self.n_components):
...: res += self.weights[i_component] * norm.cdf(x,
...: loc=self.mus[i_component],
...: scale=self.sigmas[i_component])
...: return res
...: def ppf(self, x):
...: bracket = [-10., 10.]
...: if x.min() < self.cdf(bracket[0:1]):
...: bracket[0] -= 50.
...: if x.max() > self.cdf(bracket[1:2]):
...: bracket[1] += 50.
...: res = np.array([root_scalar(lambda yy: self.cdf(np.array([[yy]]).T) - x[i],
...: bracket=bracket,
...: method='brentq',
...: xtol=1e-12, rtol=1e-12).root for i in range(len(x))])
...: return res
...:
In [6]: n_vars = 5
In [7]: margins = [None]*n_vars
In [8]: for i_var in range(n_vars):
...: n_components = 3
...: weights = np.full(n_components, 1./n_components)
...: mus = norm.rvs(size=n_components, scale=2.)
...: sigmas = np.sqrt(invwishart.rvs(df=3, scale=1.0, size=n_components))
...: margins[i_var] = GaussianMixture(weights, mus, sigmas)
...:
In [9]: cov_mat = toeplitz([np.power(0.5, k) for k in range(5)])
In [10]: u_train = norm.cdf(multivariate_normal(mean=np.zeros(5), cov=cov_mat).rvs(1000))
In [11]: x_train = np.full_like(u_train, np.nan)
In [12]: for i_var in range(n_vars):
....: x_train[:, i_var] = margins[i_var].ppf(u_train[:, i_var])
....:
In [13]: u_test = norm.cdf(multivariate_normal(mean=np.zeros(5), cov=cov_mat).rvs(1000))
In [14]: x_test = np.full_like(u_test, np.nan)
In [15]: for i_var in range(n_vars):
....: x_test[:, i_var] = margins[i_var].ppf(u_test[:, i_var])
....:
Gaussian knockoffs#
Estimate a Gaussian knockoff model
In [16]: from vineknockoffs.vine_knockoffs import VineKnockoffs
In [17]: gau_ko = VineKnockoffs()
In [18]: gau_ko.fit_gaussian_knockoffs(x_train=x_train)
Out[18]: <vineknockoffs.vine_knockoffs.VineKnockoffs at 0x7f13502b9a30>
In [19]: gau_ko._dvine.copulas
Out[19]:
[[GaussianCopula(par=[0.4489001]),
GaussianCopula(par=[0.48167758]),
GaussianCopula(par=[0.50485079]),
GaussianCopula(par=[0.51236845]),
GaussianCopula(par=[0.08826382]),
GaussianCopula(par=[0.4489001]),
GaussianCopula(par=[0.48167758]),
GaussianCopula(par=[0.50485079]),
GaussianCopula(par=[0.51236845])],
[GaussianCopula(par=[0.02854314]),
GaussianCopula(par=[-0.00969086]),
GaussianCopula(par=[0.01166561]),
GaussianCopula(par=[0.14039401]),
GaussianCopula(par=[0.07991839]),
GaussianCopula(par=[0.02854314]),
GaussianCopula(par=[-0.00969086]),
GaussianCopula(par=[0.01166561])],
[GaussianCopula(par=[0.06233617]),
GaussianCopula(par=[-0.01899161]),
GaussianCopula(par=[0.18216914]),
GaussianCopula(par=[0.16629839]),
GaussianCopula(par=[0.24476181]),
GaussianCopula(par=[0.06233617]),
GaussianCopula(par=[-0.01899161])],
[GaussianCopula(par=[0.01043043]),
GaussianCopula(par=[0.39365313]),
GaussianCopula(par=[0.39891868]),
GaussianCopula(par=[0.40050723]),
GaussianCopula(par=[0.45250961]),
GaussianCopula(par=[0.01043043])],
[GaussianCopula(par=[-0.25828746]),
GaussianCopula(par=[-0.22723834]),
GaussianCopula(par=[-0.08053081]),
GaussianCopula(par=[-0.07758852]),
GaussianCopula(par=[-0.35665741])],
[GaussianCopula(par=[0.64614131]),
GaussianCopula(par=[0.54497431]),
GaussianCopula(par=[0.46927182]),
GaussianCopula(par=[0.71024124])],
[GaussianCopula(par=[-0.55794148]),
GaussianCopula(par=[-0.38227897]),
GaussianCopula(par=[-0.48864249])],
[GaussianCopula(par=[0.64844621]), GaussianCopula(par=[0.41442168])],
[GaussianCopula(par=[-0.99475601])]]
Generate a knockoff copy
In [20]: x_gau_ko = gau_ko.generate(x_test)
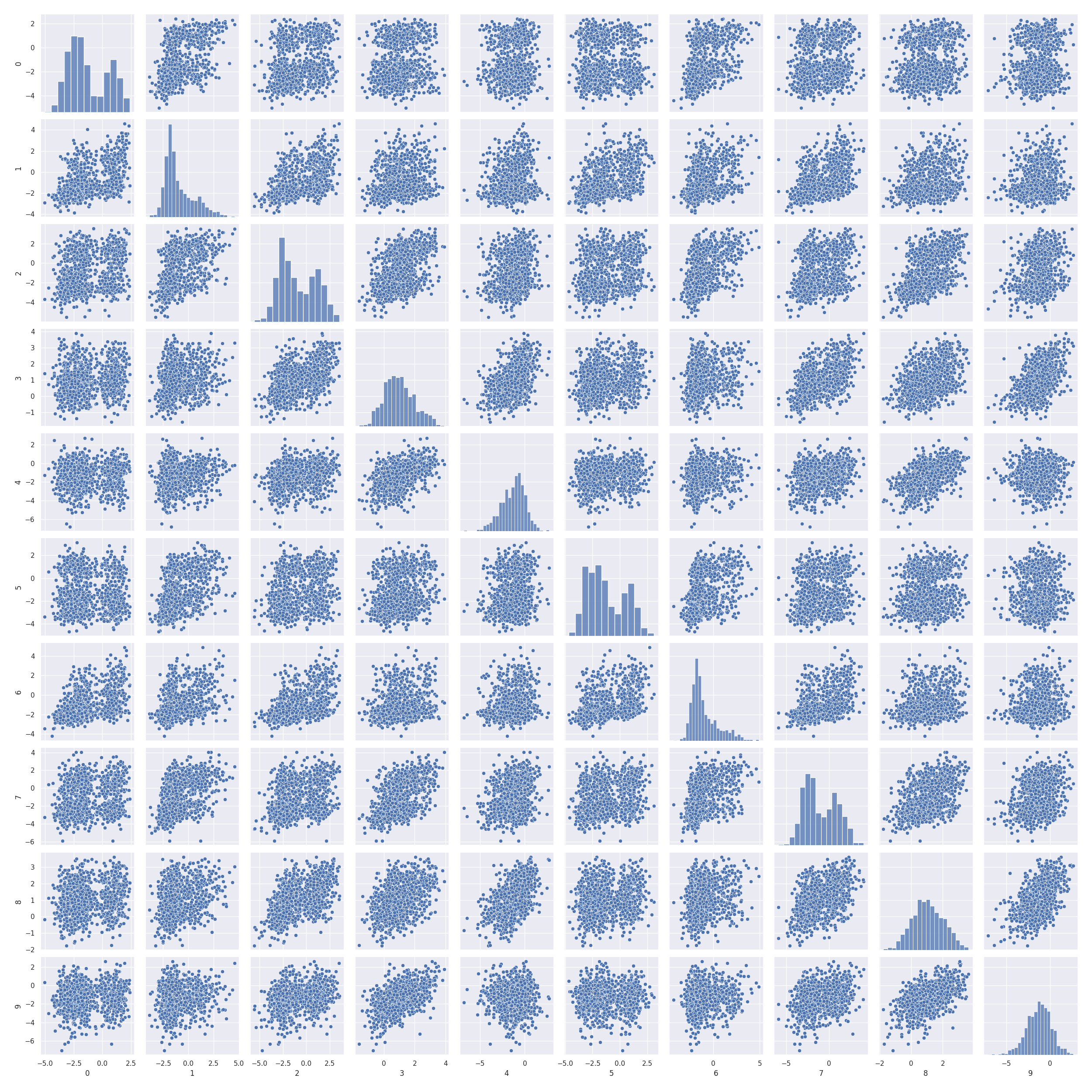
Pairwise scatter plots
In [21]: import pandas as pd
In [22]: import seaborn as sns
In [23]: import matplotlib.pyplot as plt
In [24]: sns.set(font_scale=1)
In [25]: sns.pairplot(pd.DataFrame(np.hstack((x_test, x_gau_ko))))
Out[25]: <seaborn.axisgrid.PairGrid at 0x7f1348f55cd0>

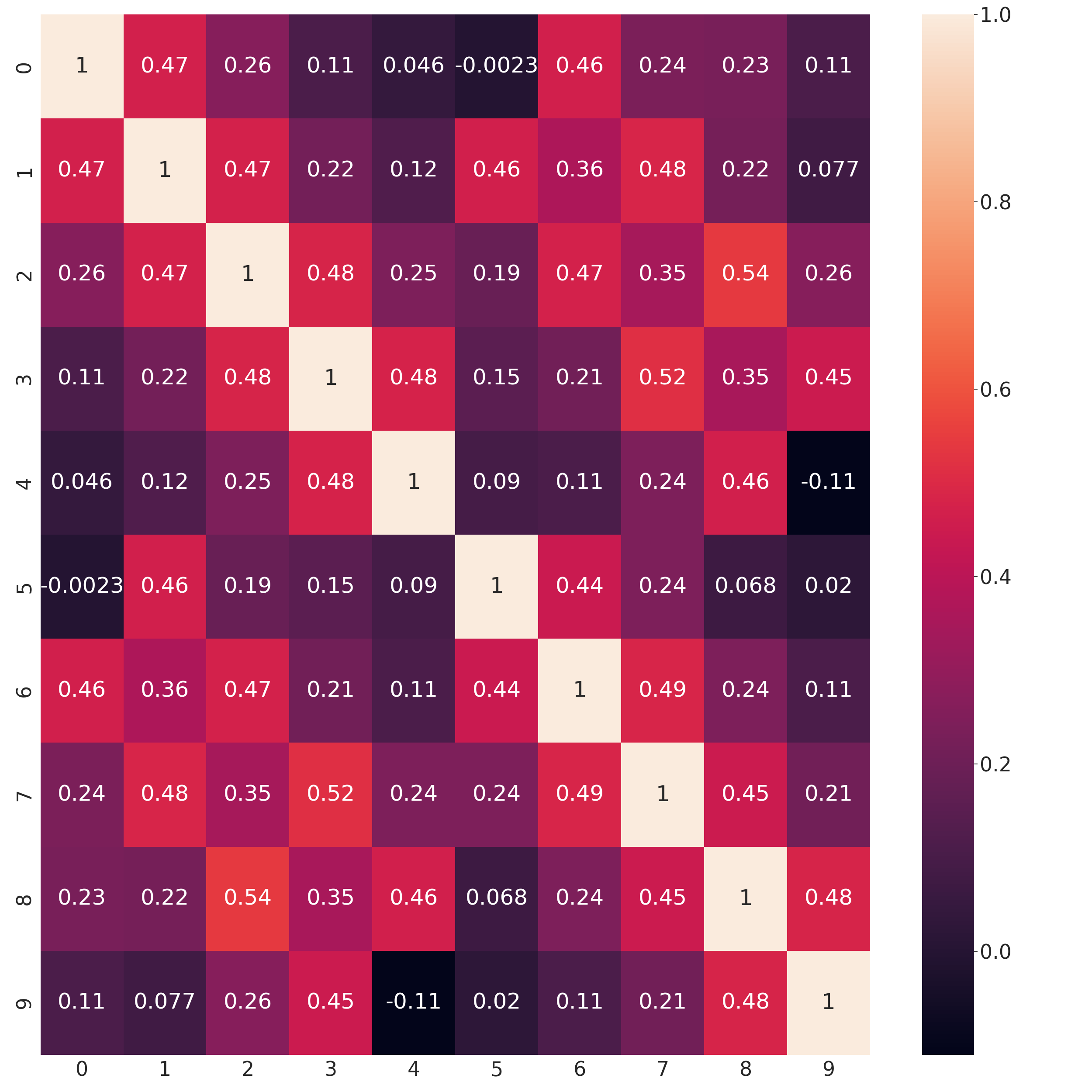
Estimated correlations
In [26]: sns.set(font_scale=3)
In [27]: sns.heatmap(np.corrcoef(np.hstack((x_test, x_gau_ko)).T), annot=True);

Gaussian copula knockoffs#
Estimate a Gaussian copula knockoff model
In [28]: from vineknockoffs.vine_knockoffs import VineKnockoffs
In [29]: gau_cop_ko = VineKnockoffs()
In [30]: gau_cop_ko.fit_gaussian_copula_knockoffs(x_train=x_train)
Out[30]: <vineknockoffs.vine_knockoffs.VineKnockoffs at 0x7f1347498760>
In [31]: gau_cop_ko._dvine.copulas
Out[31]:
[[GaussianCopula(par=[0.49889927]),
GaussianCopula(par=[0.51485352]),
GaussianCopula(par=[0.51563259]),
GaussianCopula(par=[0.51825192]),
GaussianCopula(par=[0.08920866]),
GaussianCopula(par=[0.49889927]),
GaussianCopula(par=[0.51485352]),
GaussianCopula(par=[0.51563259]),
GaussianCopula(par=[0.51825192])],
[GaussianCopula(par=[-0.00416141]),
GaussianCopula(par=[-0.01605925]),
GaussianCopula(par=[0.00478767]),
GaussianCopula(par=[0.15206381]),
GaussianCopula(par=[0.09216385]),
GaussianCopula(par=[-0.00416141]),
GaussianCopula(par=[-0.01605925]),
GaussianCopula(par=[0.00478767])],
[GaussianCopula(par=[0.06845161]),
GaussianCopula(par=[-0.01260832]),
GaussianCopula(par=[0.19342715]),
GaussianCopula(par=[0.17230366]),
GaussianCopula(par=[0.24343657]),
GaussianCopula(par=[0.06845161]),
GaussianCopula(par=[-0.01260832])],
[GaussianCopula(par=[0.00267909]),
GaussianCopula(par=[0.44566922]),
GaussianCopula(par=[0.43046756]),
GaussianCopula(par=[0.4090201]),
GaussianCopula(par=[0.45793907]),
GaussianCopula(par=[0.00267909])],
[GaussianCopula(par=[-0.33754818]),
GaussianCopula(par=[-0.00999287]),
GaussianCopula(par=[-0.11597554]),
GaussianCopula(par=[-0.0109706]),
GaussianCopula(par=[-0.36733475])],
[GaussianCopula(par=[0.63963203]),
GaussianCopula(par=[0.48851243]),
GaussianCopula(par=[0.46462707]),
GaussianCopula(par=[0.68064834])],
[GaussianCopula(par=[-0.5505026]),
GaussianCopula(par=[-0.33889836]),
GaussianCopula(par=[-0.46158122])],
[GaussianCopula(par=[0.63887789]), GaussianCopula(par=[0.37102548])],
[GaussianCopula(par=[-0.99548401])]]
Generate a knockoff copy
In [32]: x_gau_cop_ko = gau_cop_ko.generate(x_test)
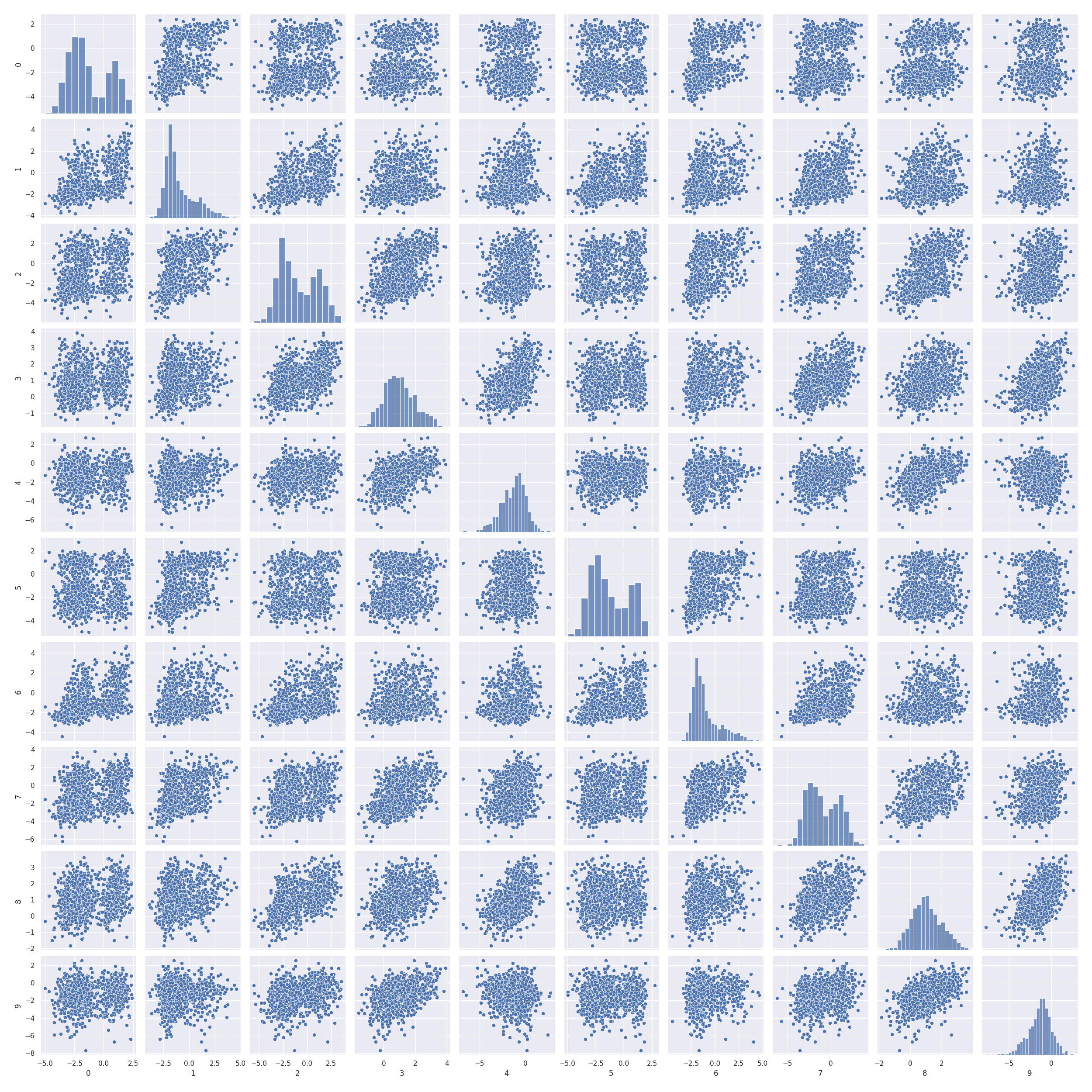
Pairwise scatter plots
In [33]: import pandas as pd
In [34]: import seaborn as sns
In [35]: import matplotlib.pyplot as plt
In [36]: sns.set(font_scale=1)
In [37]: sns.pairplot(pd.DataFrame(np.hstack((x_test, x_gau_cop_ko))))
Out[37]: <seaborn.axisgrid.PairGrid at 0x7f1347b128e0>
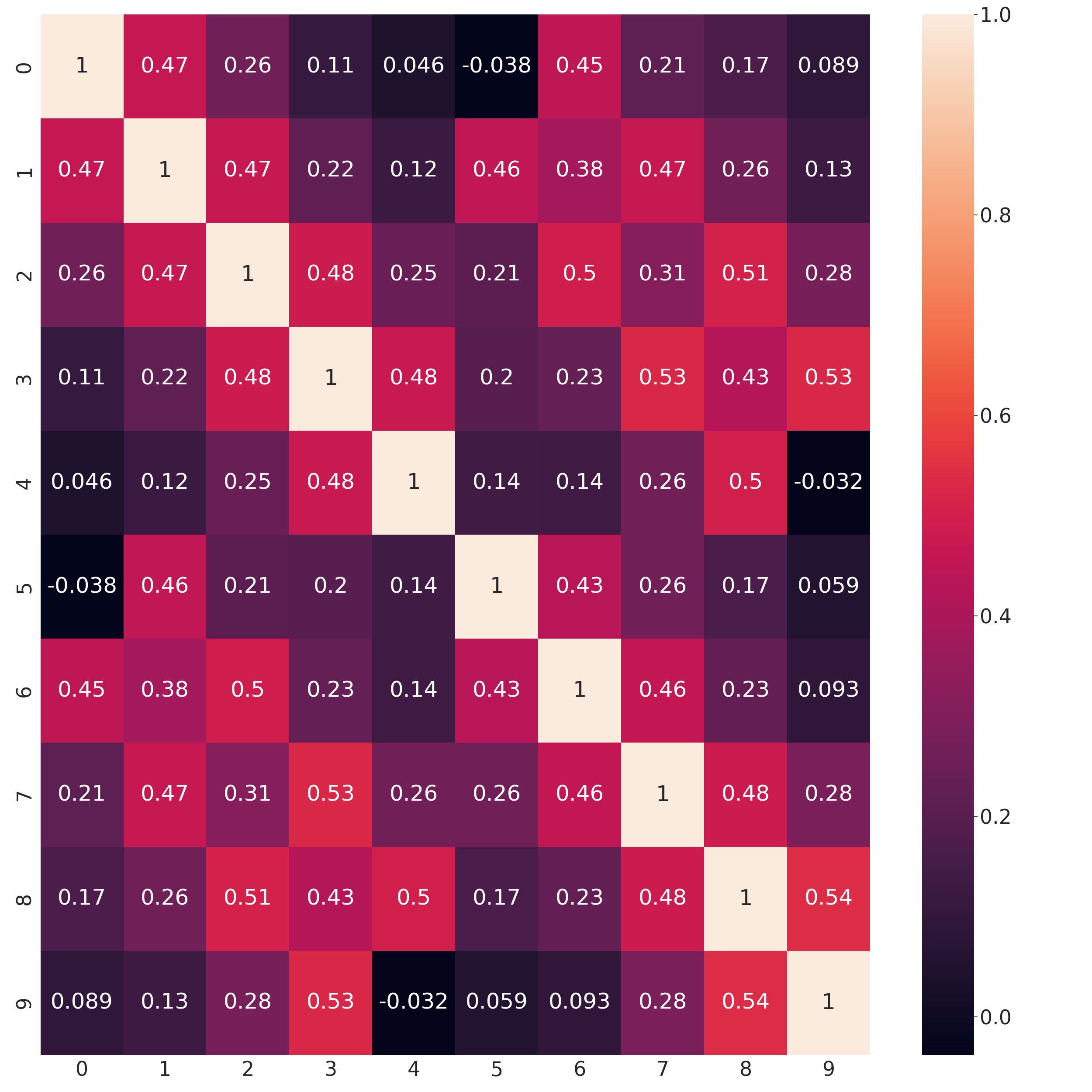
Estimated correlations
In [38]: sns.set(font_scale=3)
In [39]: sns.heatmap(np.corrcoef(np.hstack((x_test, x_gau_cop_ko)).T), annot=True);
Vine copula knockoffs#
Estimate a Gaussian copula knockoff model
In [40]: from vineknockoffs.vine_knockoffs import VineKnockoffs
In [41]: vine_ko = VineKnockoffs()
In [42]: vine_ko.fit_vine_copula_knockoffs(x_train=x_train)
Out[42]: <vineknockoffs.vine_knockoffs.VineKnockoffs at 0x7f13447aa490>
In [43]: vine_ko._dvine.copulas
Out[43]:
[[GaussianCopula(par=[0.48383016]),
GaussianCopula(par=[0.51726826]),
GaussianCopula(par=[0.27181872]),
GaussianCopula(par=[0.51290159]),
GaussianCopula(par=[0.16647727]),
GaussianCopula(par=[0.48383016]),
GaussianCopula(par=[0.51726826]),
GaussianCopula(par=[0.27181872]),
GaussianCopula(par=[0.51290159])],
[IndepCopula(),
IndepCopula(),
GaussianCopula(par=[0.45121434]),
IndepCopula(),
GaussianCopula(par=[0.19896423]),
IndepCopula(),
IndepCopula(),
GaussianCopula(par=[0.45121434])],
[IndepCopula(),
IndepCopula(),
ClaytonCopula(par=[0.22514369], rotation=180),
IndepCopula(),
GaussianCopula(par=[0.46917166]),
IndepCopula(),
IndepCopula()],
[IndepCopula(),
GaussianCopula(par=[0.44616686]),
GaussianCopula(par=[0.43643683]),
IndepCopula(),
GaussianCopula(par=[0.45584443]),
IndepCopula()],
[GaussianCopula(par=[-0.33754818]),
GaussianCopula(par=[-0.00999287]),
GaussianCopula(par=[-0.11597554]),
GaussianCopula(par=[-0.36733475]),
GaussianCopula(par=[-0.0109706])],
[GaussianCopula(par=[0.63963203]),
GaussianCopula(par=[0.48851243]),
GaussianCopula(par=[0.01680553]),
GaussianCopula(par=[0.68064834])],
[GaussianCopula(par=[-0.5505026]),
GaussianCopula(par=[-0.03065251]),
GaussianCopula(par=[0.61868149])],
[GaussianCopula(par=[0.0467642]), GaussianCopula(par=[-0.48578146])],
[GaussianCopula(par=[0.99732389])]]
Generate a knockoff copy
In [44]: x_vine_ko = vine_ko.generate(x_test)
Pairwise scatter plots
In [45]: import pandas as pd
In [46]: import seaborn as sns
In [47]: import matplotlib.pyplot as plt
In [48]: sns.set(font_scale=1)
In [49]: sns.pairplot(pd.DataFrame(np.hstack((x_test, x_vine_ko))))
Out[49]: <seaborn.axisgrid.PairGrid at 0x7f1344bc5df0>
Estimated correlations
In [50]: sns.set(font_scale=3)
In [51]: sns.heatmap(np.corrcoef(np.hstack((x_test, x_vine_ko)).T), annot=True);